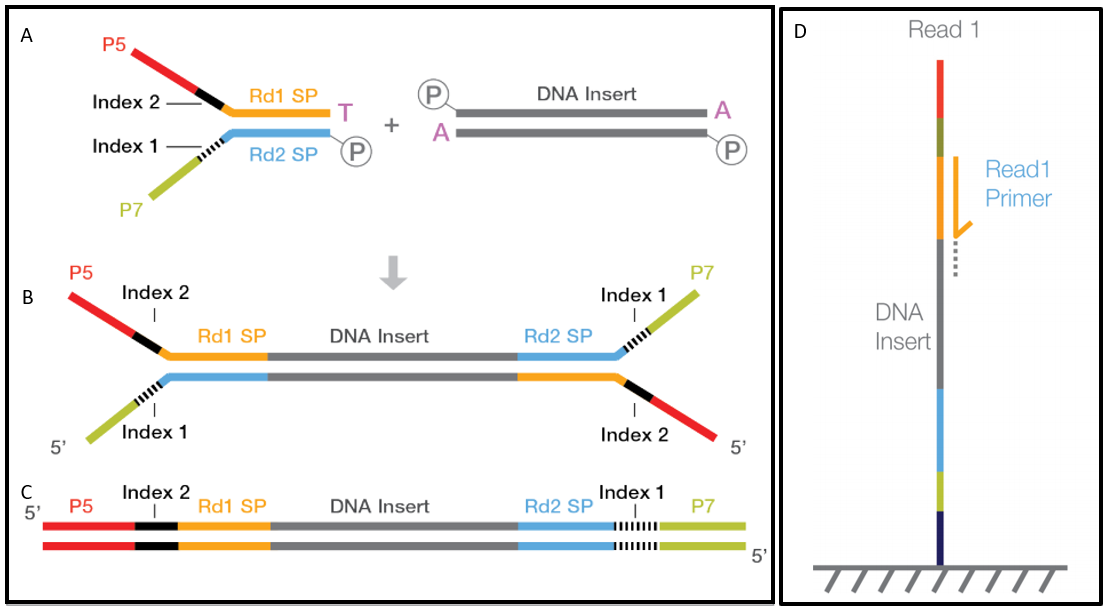
Figure 1 sequencing library after paired‐end sample preparation the two adapters contain sequences that are complimentary to the two surface‐bound. If nominal length is 500bp, put down 500 as the value.

Sequencing Library Information Submission Guide Annotare Embl-ebi
The insert being the fragment sequenced, as chosen from size fractionation (e.g.

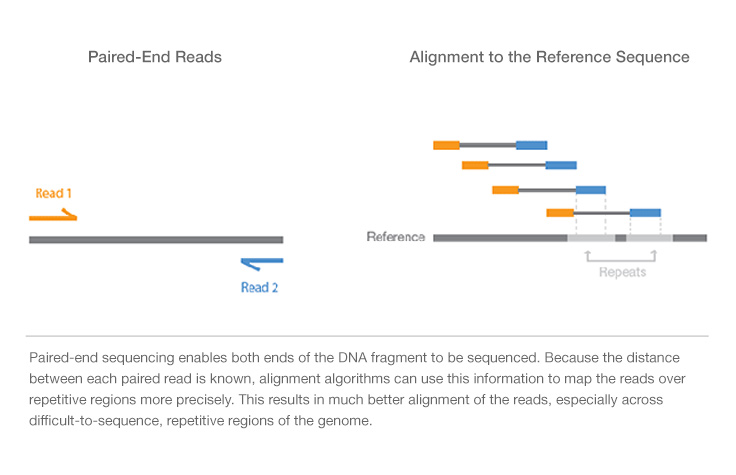
Paired end sequencing insert size. For instance, for a pe150 sequencing run, the insert size should be above 300bp. The collectinsertsizemetrics tool outputs the percentages of read pairs in each of the three orientations (fr, rf, and tandem) as a histogram. A selection of 29 lbs (additional file 1:
We aimed at obtaining at least 80 million reads per sample, employing the illumina novaseq 6000 (illumina, essex, uk). The distribution is somewhat leptokurtic and positively skewed with a minimum insert size around 40 bp and maximum insert size around 850 bp. The variation of insert sizes is often large and the average size difficult to control.
So why you get reads of the same length when. The purpose of this protocol is to add adapter sequences onto the ends of dna fragments to generate the following sequencing library format: In the answer, it is mentioned that the insert size is the library size minus your two read pairs' sizes.
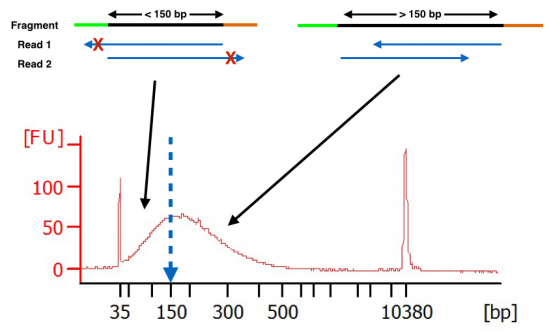
All paired read data will have a known expected distance between each pair given the experimental design. Normally the insert size is longer than the sum of the two read lengths, meaning there is an unsequenced inner part in the middle of the insert. This can result in a proportion of fragments with an insert size of less than the length of a single read.
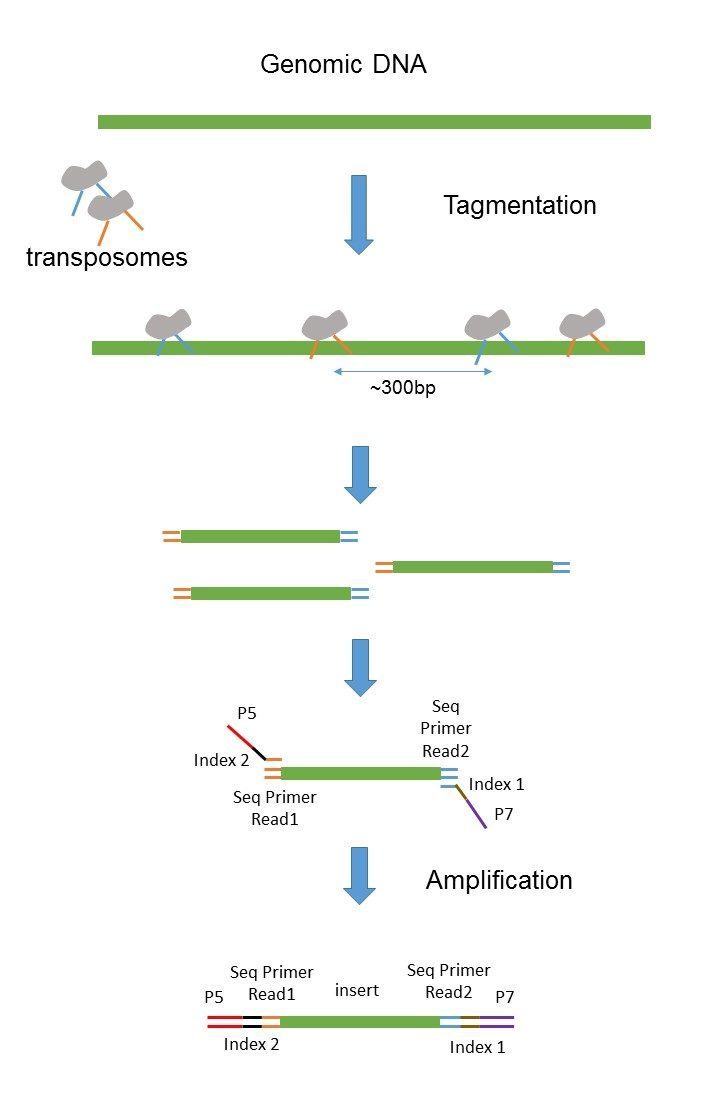
Set output format as sam(bowtie) or bam(bwa). The sequencing starts at read 1 adapter (mate 1) and ends with the sequencing from read 2 adapter (mate 2). Size‐selected dna is pcr amplified to enrich for.
This area that is not sequenced between the read pairs. i am afraid that this is not. In addition, the insert size distribution is output as both a histogram (.insert_size_histogram.pdf) and as a data table (.insert_size_metrics.txt). The fragment size (which you need to select for during a gel purification for example) would be the insert size + length of both adapters (around 120 bp extra for both illumina adapters).
It is important you set this to the correct value to achieve good results when assembling. Including flanking intronic regions of diagnostic significance (e.g., due to splice variants) of about 30bp at either side,. Cutting out a band from an agarose gel).
However, the average size of human coding exons is only 160bp. No decimals or ranges (e.g. Typically illumina paired end reads have an insert longer than the combined length of both reads (see figure figure1 1).
The expected size of the insert. Using the latest illumina platform, the miseq, paired end reads of 250 (and recently even.

Five Approaches To Detect Cnvs From Ngs Short Reads A Paired-end Download Scientific Diagram
What Happens When Hardware Advances Faster Than Molecular Protocols Cofactor Genomics
How Short Inserts Affect Sequencing Performance

What Is Mate Pair Sequencing For
What Is Paired-end 150 Omega Bioservices
Ngs

Interpreting Color By Insert Size Integrative Genomics Viewer
The Insert-size In Paired-end Data - Seqanswers

What Are Paired-end Reads The Sequencing Center
What Read Lengths Are Produced By Modern Illumina Sequencers

Definition Of Essential Terms A Definition Of Fragment Read And Download Scientific Diagram
Paired-end Sequencing - France Genomique
Paired End Illumina Reads

Diagram To Show The Construction Of A Fragment With An Insert Size Is Download Scientific Diagram

Interpreting Color By Insert Size Integrative Genomics Viewer

What Is Mate Pair Sequencing For

Detecting Deletion Events A When Mapping Paired-end Reads To The Download Scientific Diagram

Library Insert Size Libraries Were Constructed Using The Standard A Download Scientific Diagram

Pdf Assessment Of Insert Sizes And Adapter Content In Fastq Data From Nexteraxt Libraries
Paired End Sequencing Insert Size. There are any Paired End Sequencing Insert Size in here.